StressME
Project Details
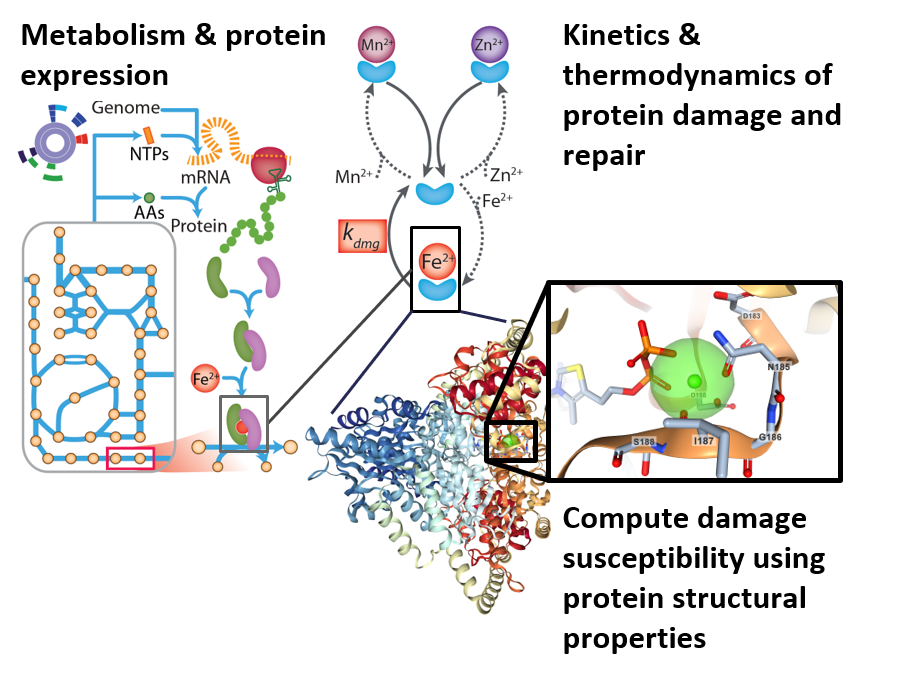
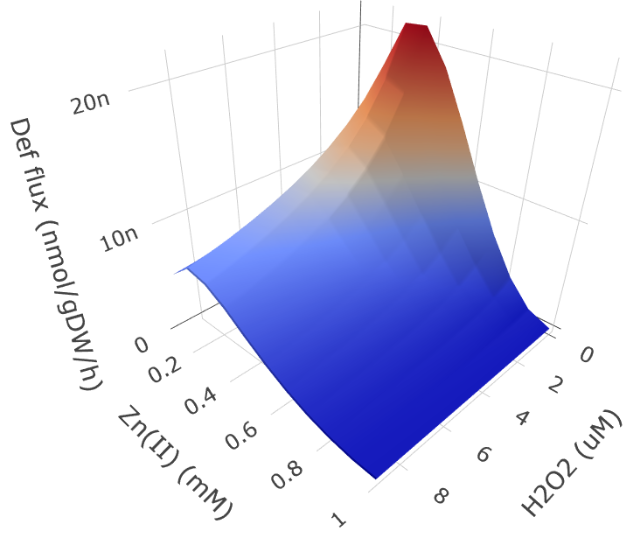
Aerobic organisms have evolved cellular responses against reactive oxygen species (ROS) over the coures of 3 billion years, since the Earth's atmosphere became oxygenated. ROS targets macromolecules and metal cofactors, which can impair cellular function. ROS is generated as a by-product of normal metabolic operation. It is also used as an offensive tool by competing microbes and the human immune system.
Adaptation against ROS stress can require system-level responses, including metabolic shifts, use of alternative cofactors, and change in macromolecular composition. Adaptation against other stresses (e.g., thermal, pH, osmotic) requires similar system level responses.
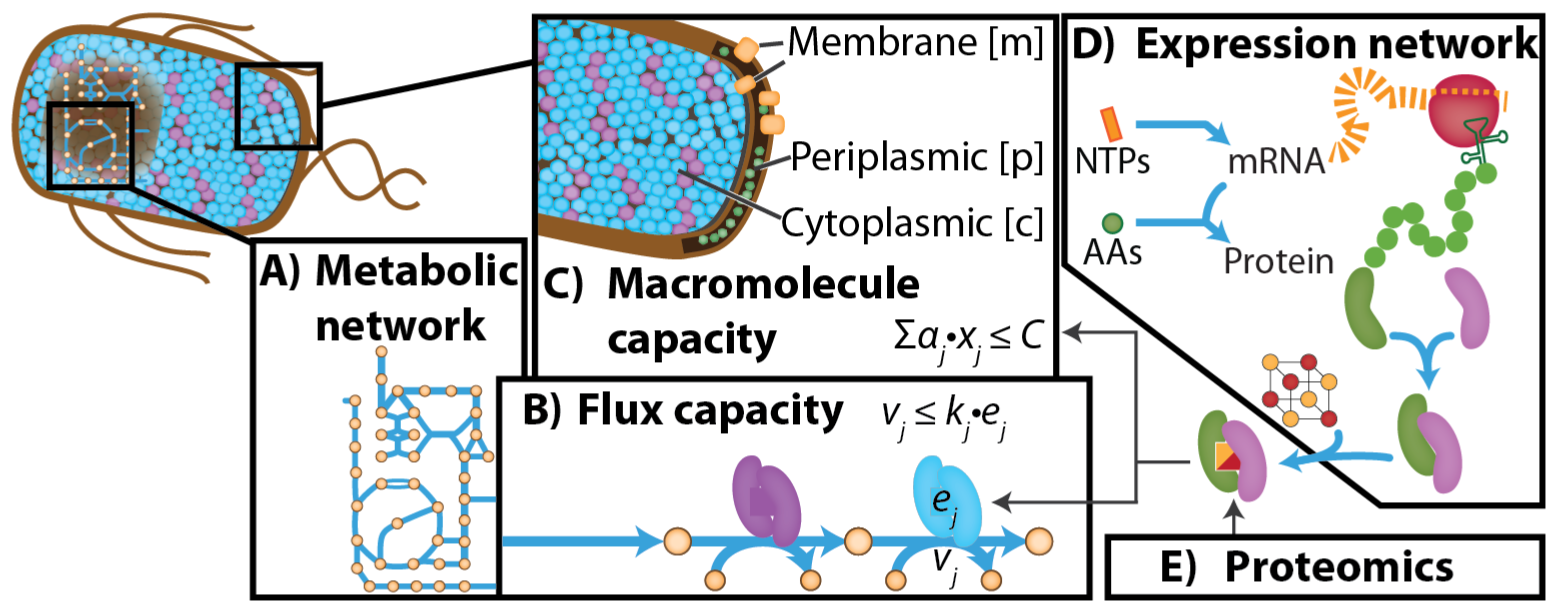
Such system-level stress responses can be modeled using genome-scale models that integrate the processes of metabolism and macromolecule expression (ME models). By extending these models with the thermodynamic and kinetic processes associated with stress-related damage and repair of cellular components, it is possible to model microbial stress response. These extended models are collectively termed StressME models.
Our lab aims to:
- model additional key stress responses (e.g., osmotic stress)
- investigate the role of stress tolerance in infectious disease
- model cellular response to osmotic, oxidative, and pH stress in bioprocess environments
Researchers

Laurence Yang
Assistant Professor

Jiao Zhao
Research Associate

Sanjeev Dahal
Postdoctoral Fellow

Hao Xu
Research Assistant
- Publications
- FoldME paper
- OxidizeME paper
- AcidifyME paper
- Synthesizing systems biology knowledge from omics using genome-scale models
- StressME paper (coming soon)
- Software
- OxidizeME code
- AcidifyME code
- StressME app (coming soon)
- Funding
- 2020-2022: subaward of NIH Grant R01GM057089 (PI: Bernhard O. Palsson, UCSD)